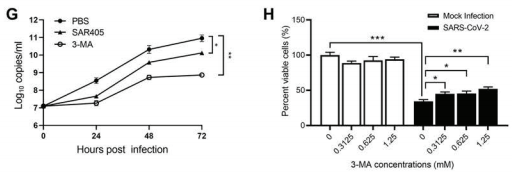
Colorectal cancer is the third most diagnosed cancer and leads to the second mortality among cancers worldwide. The first-line chemotherapeutic drugs of colorectal cancer are mainly 5-fluorouracil(5-FU) based. However, these drugs exhibit compromised efficacy due to significant toxicity, drug resistance or patient inconvenience. Novel chemotherapeutic drugs for efficient treatment of colorectal cancer patients are urgently needed. Cabazitaxel (CBT), a microtuble inhibitor, can promotes polymerization of tubulin and stabilize microtubules. It shows antitumor activity in docetaxel-refractory metastatic prostate cancer and breast cancer.
However, it is not clear whether CBT is effective in inhibiting colorectal cancer and what the underlying mechanism is. In response to this question, The researchers from The Sixth Affiliated Hospital of Sun Yat-sen University, Wen Zhang et al. published an article named “Cabazitaxel suppresses colorectal cancer cell growth via enhancing the p53 anti-tumor pathway” in FEBS Open Bio. In this study, the researchers screened 160 FDA approved drugs with HCT116 cells, the commonly used colorectal cancer cell lines, and found that Cabazitaxel, by enhancing the P53 antitumor pathway, can efficiently inhibit the proliferation and migration of colorectal cancer cells via a series of in vitro assays.

Cabazitaxel inhibits colorectal cancer cell growth via activating p53 signaling pathway
KEGG pathway analysis revealed that CBT treatment induced genes were enriched in the well-known anti-tumor p53 signaling pathway (Fig. 1). Further gene set enrichment analysis (GSEA) revealed a positive correlation between p53 pathway genes with CBT upregulated genes in HCT116 cells, indicating that CBT indeed enhances the expression of p53 pathway genes (Fig. 2). To examine whether activation of p53 pathway plays a key role for CBT efficacy, they used CRISPR/Cas9 system to knock out TP53, p53 encoding gene, in HCT116 cells by two different gRNAs and generate TP53 KO1 cells and TP53 KO2 cells with two hTP53 gRNA KO plasmids (YKO-RP003-hTP53, YKO-RP003-hTP53) which were provided by Ubigene. MTT assay revealed that IC50 of CBT to TP53 KO1 cells and TP53 KO2 cells were about at least three fold higher than IC50 of CBT in HCT116 cells. The enhanced resistance to CBT of TP53 KO cells indicates that the inhibitory effect of CBT to HCT116 cells relies on the TP53 pathway (Fig. 3). All these results substantiate that CBT inhibits HCT116 cells mainly via activating p53 pathway.
In this study, Ubigene provided hTP53 gRNA KO plasmids for the construction of TP53 knockout cell line. It was demonstrated that CBT inhibited HCT116 cells mainly by activating p53 pathway. Ubigene now provides TP53 knockout cell lines, which can be delivered within one week for only $1980. Click to learn more > >
If gRNA plasmid is needed, welcome to visit our gRNA plasmid bank for retrieval. Click for more details > >

Fig. 1
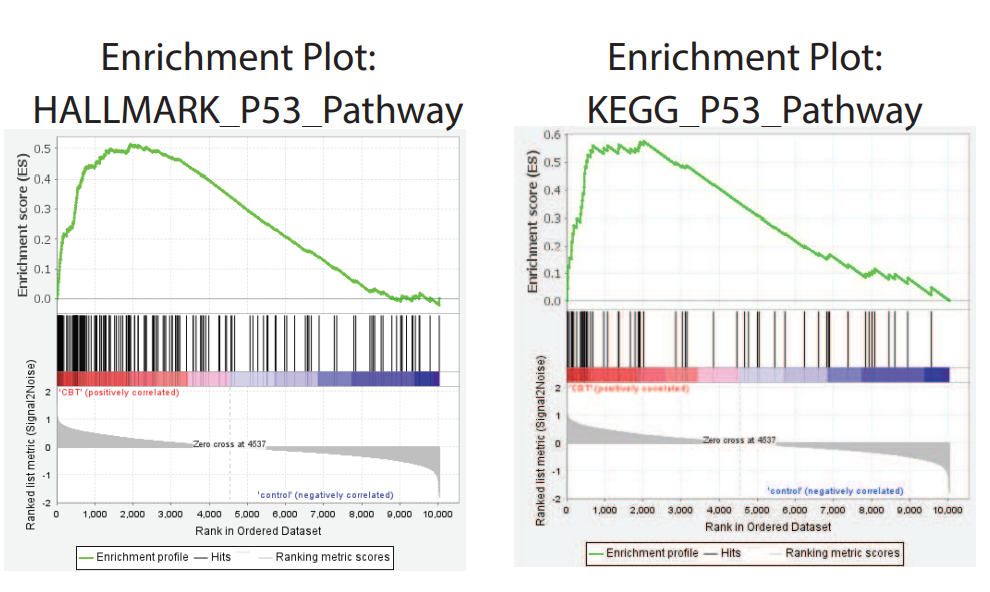
Fig. 2
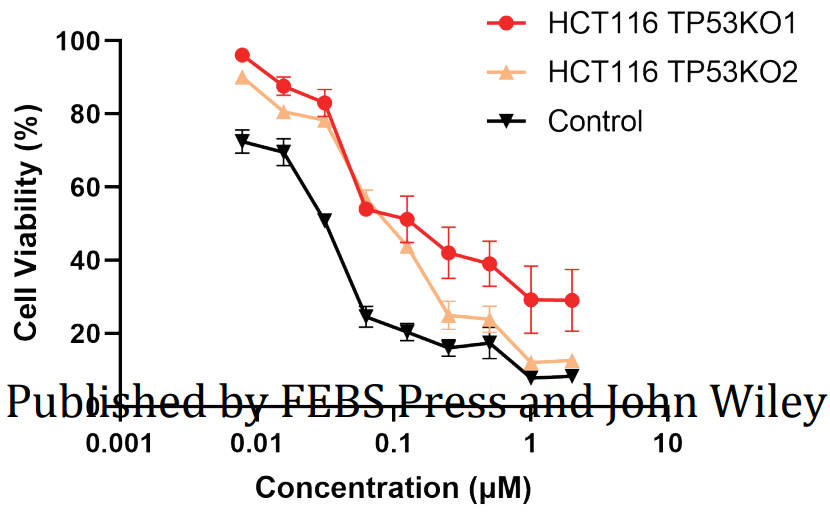
Fig. 3
In this study, they showed that CBT can efficiently inhibit colorectal cancer proliferation and migration. It suppresses colorectal cancer via enhancing the expression of multiple p53 downstream effector genes and promoting cell cycle arrest, apoptosis and inhibition of angiogenesis. Hence, CBT may be served as an alternative option for colorectal cancer treatment in future.
Ubigene is mainly focusing on cell engineering. We provide high-quality gene-editing cell lines, stable cell lines, virus packaging and other related services around the world, as well as nearly 2000 KO cell bank, gene knockout kits and other gene-editing related products.
More articles:
Well-known SW480 cell line is not “simple”
Gene editing in BEAS-2B cell line facilitates respiratory researches.