Background
293T is a popular cell line for proof of concept. It can express SV40 T-antigen and contain SV40 origin of replication and promoter region. Many eukaryotic expression plasmids containing SV40 virus origin of replication, such as pcDNA3.1, can replicate in 293T cell line, so 293T cell line is widely used in virus packaging. 293T cell line also can be used for studying the expression of exogenous genes. Firstly, 293T cell line is easy to be transfected, and the transfection efficiency can be as high as 50% with calcium phosphate; Secondly, the protein expression level of 293T is high. The expressed protein can be easily detected by alkaline phosphatase analysis in 2-3 days after transfection. In addition, the transfection of the 293T cell line is one of the convenient ways to overexpress proteins and obtain intracellular and extracellular proteins. Thus, the 293T cell line has provided great help for a large number of cells and molecular level-related research. If you need to purchase 293T cells which have low passages, high activity and good cell condition, please click here >>
Detailed applications
293T cell line is widely used for researches, such as Lentivirus packaging and titer test, gene expression research, protein expression research, signaling pathway research, antibody screening, antibiotic screening, conditioned culture medium production, etc. Thus, 293T cell line is considered basic cell line by the researchers.
Applications of CRISPR/Cas9 technology in 293T cell line
CRISPR/Cas9 technology, as a popular gene-editing technology at present, has been well applied in 293T cell line. The stable cell lines are constructed through accurate gene-editing, which can be used to study apoptosis, functional genomics, signaling pathways, drug R&D, drug resistance mechanisms, drug response, and cell therapy. Ubigene’s developed CRISPR-U™ technology is 10-20 times more efficient than traditional CRISPR/Cas9 technology, and can greatly improve the recombination efficiency, which provides a powerful tool for gene-editing in 293T cell line.
So far, Ubigene has successfully generated a large number of gene KO 293T cell lines, the in-stock KO cell line is only 1880 USD. And we plan to construct ~900 genes knockout cell lines on the 293T cell line. These genes come from 8 popular signaling pathways and have high research value. If you are interested in our in-stock KO cell lines, please feel free to contact us!
Specific cases of gene-editing in 293T cell line
1. 293T cell line plays a key role in studying the effect of PLAC8 on the proliferation and migration of kidney cancer cells
The oncogene placental specific 8 (PLAC8) plays an important role in cellular processes and human diseases, but there are few studies on how PLAC8 affects the proliferation and migration of Human kidney cancer cells (KC). Therefore, Qin et al. selected 293T cell line as the target cell line to study the effect of PLAC8 on the proliferation and migration of 293T cell line. Firstly, they established two PLAC8 KO 293T clones with CRISPR/Cas9. In order to classify the characteristics of PLAC8 in the process of 293T cell line proliferation and migration, they conducted cell phenotypic tests such as cell counting, colony formation assay(cell proliferation), cell cycle, cell scratch test, and cell migration/Invasion(Ubigene provides various Cell Phenotypic Services). The molecular mechanism was studied by Western blot. The results showed that KO PLAC8 could inhibit the proliferation of the 293T cell line. In addition, the inhibitory effect of PLAC8 KO on 293T cell line proliferation was related to G2/M in the cell cycle. At the same time, PLAC8 KO significantly inhibited cyclin B1 and increased cyclin a. This shows that PLAC8 plays an important role in 293T cell line proliferation and migration, and provides an important research basis for further study of the effect of PLAC8 on human KC cells [2].
2. 293T cell line helps to build a quantitative model of recombinant protein production
293T cell line can express foreign genes. So it has been considered a powerful tool to produce and express specific protein products, and has been widely used to produce a variety of recombinant proteins. However, the recombinant protein yield of constructed cell lines is often inconsistent and difficult to quantify. Lo et al. used CRISPR/Cas9 to bind human ribosomal protein L13A(RPL13A) KI to a specific site (Fig. 3), so that transgene expression is driven by endogenous promoters to ensure consistent and predictable expression of recombinant proteins. The expression level can also be determined in advance by selecting promoters from genes with the required expression level. In order to quantify the expression of recombinant protein, protein quantitative reporter gene (PQR) is integrated between endogenous gene and exogenous gene. PQR equimolar produces endogenous protein, recombinant protein and fluorescence reporter gene, so the expression of target protein can be quantified by cell fluorescence intensity.[3]
Ubigene specialized in gene KI cell line generation, as low as 9480 USD, it will come with a $2000 coupon for cell phenotypic services. For more details, please inquire >>
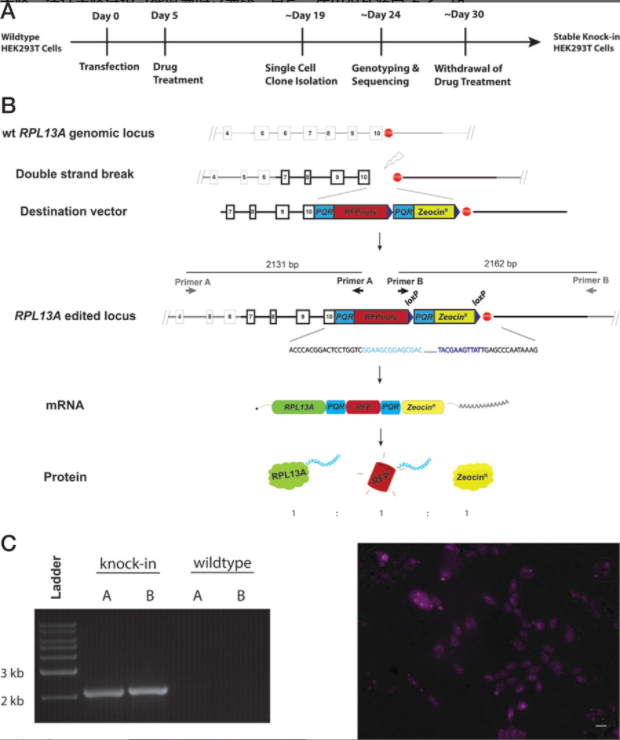
Figure 1 RPL13A KI 293T cell line generation
3. Study of telomerase activity inhibition by 293T cell line
All continuously proliferating cell lines, such as stem cells and cancer cells, have a mechanism to compensate for telomere wear during continuous division. In most cases, this requirement is met by telomerase. Somatic cell line cannot divide indefinitely due to the lack of telomerase activity, but the reactivation of TERT transcription in cell lines will enable them to continue to divide, which is a key step in the process of tumorigenesis. Therefore, the study of TERT expression is of great significance to understand the regulatory mechanism of telomerase activity under physiological and pathological conditions. Two point mutations in the promoter region of the human TERT gene (C-124T and C-146T) are highly recurrent in various cancer types and are associated with higher telomerase levels. Xi et al. modified the endogenous TERT gene with CRISPR/Cas9 and labeled the endogenous TERT protein or protein with localization tags. They generated a 293T cell line expressing FLAG-SNAP labeled TERT protein, which realized the effective immunopurification (IP) and subcellular localization of endogenous TERT. The results showed that telomerase was localized to only a few telomeres at any given time. They also generated HEK 293T and SCaBER cell line with a modified TERT promoter, suggesting that removing the C-124T mutation is sufficient to reduce telomerase levels and shorten telomeres. This method not only provides a useful tool for the study of telomerase biology, but also provides a general method to purify and visualize low abundance proteins and modify single base pairs at genomic sites with low editing efficiency.[4]
Based on over 5000 successful gene-editing cases, Ubigene provides high-quality gene-editing cell line services, including knockout, knockin, and point mutation. We also offer stable cell line generation service for customers. If you are interested in one of our services, please feel free to contact us!