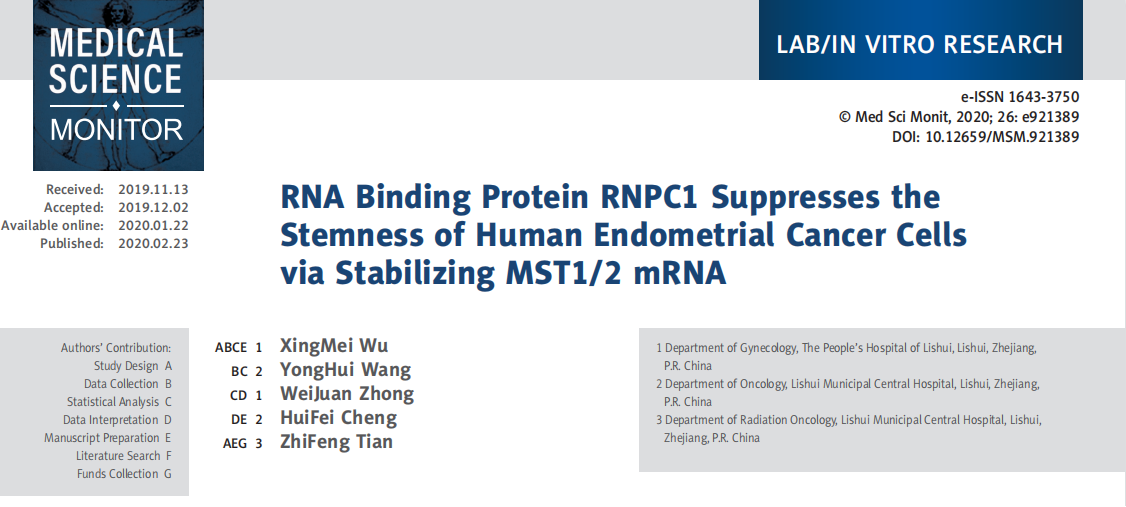
Nonalcoholic fatty liver disease (NAFLD) is a major liver disease subtype , which is characterized by an excessive hepatic lipid accumulation, covers a wide range of liver disorders, including steatosis, steatohepatitis, fibrosis, and cirrhosis. Metformin is commonly used to treat type 2 diabetes mellitus and, in recent years, it was found to play a potential role in the amelioration of NAFLD. Transcription factor EB (TFEB) is a master transcriptional regulator of lysosomal biogenesis and autophagy and, when activated, is effective against disorders of lipid metabolism.
TFEB plays a key role in lipid metabolism. However, whether TFEB is involved in the protective effect of metformin against NAFLD has not yet been reported. In response to this question, Dan Zhan et al. from first Affiliated Hospital of Kunming Medical University published an article “Metformin Alleviates Hepatic Steatosis and Insulin Resistance in a Mouse Model of High-Fat DietInduced Nonalcoholic Fatty Liver Disease by Promoting Transcription Factor EB-Dependent Autophagy” in Frontiers in Pharmacology.
In this report, mice were divided into five groups: 1) the control group, 2) high fat diet (HFD)-fed group, 3) HFD + Metformin (Met) group, 4) HFD + Met + Scramble control group, and 5) HFD + Met + TFEB shRNA group. The result demonstrates that the activity of TFEB is reduced in the liver of mice fed a high-fat diet. Metformin treatment significantly reverses the activity of TFEB, and the protective effect of metformin against hepatic steatosis and insulin resistance is dependent on TFEB. They found that metformin-induced autophagy is regulated by TFEB, and the findings reveal that TFEB acts as a mediator, linking metformin with autophagy to reverse NAFLD, and highlight that TFEB may be a promising molecular target for the treatment of NAFLD.

Metformin Induces Autophagy in the Liver of NAFLD Mice
A metformin treatment can signifificantly improve an excessive lipid accumulation, hepatic steatosis, and IR in the mice fed an HFD. And an autophagy dysregulation may contribute to the pathogenesis of NAFLD. So the researchers hypothesized that metformin reduces obesity, hepatic steatosis, and IR, probably via the activation of autophagy. To verify the activation of autophagy in the liver, they evaluated the levels of autophagy proteins Atg7, LC3B-II, as well as autophagic substrate p62/SQSTM1 by using Western blotting. The results demonstrate that metformin promotes autophagy in the liver of HFD-induced NAFLD mice (Figure 1).
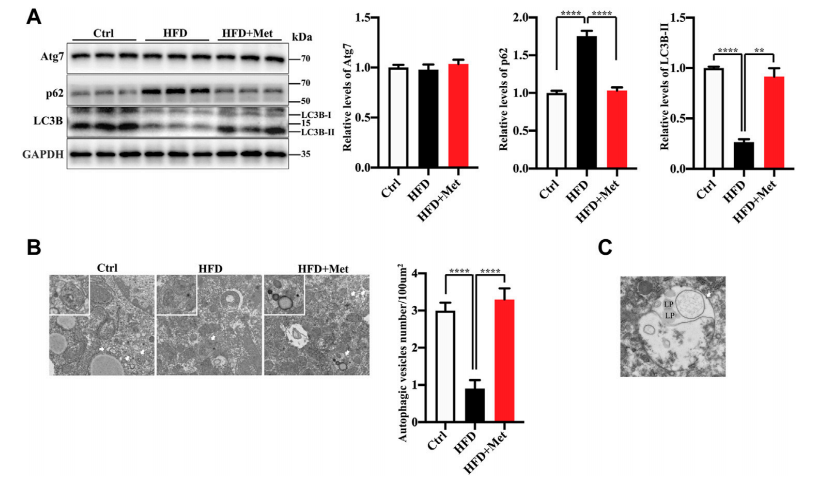
Figure 1
Metformin Induces Autophagy Through the Activation of TFEB in the Liver of NAFLD Mice
TFEB is a major regulator of autophagy and lysosomal biogenesis. The activity of TFEB mainly depends on its phosphorylation status and cytoplasm-nucleus shuttling , and the nuclear translocation is a hallmark of TFEB activation. Firstly, the researchers used Western blotting to examined whether metformin activated TFEB, the result showed that metformin treatment resulted in an increased nuclear accumulation of TFEB (Figure 2A) and reduced phosphorylation of TEFB. Then, to further test whether TFEB is involved in metformininduced autophagy, the researchers performed the knockdown of TFEB via tail vein injection with an AAV8 virus expressing an shRNA (provided by Ubigene) that targets TFEB (shTFEB). Compared with the injections of a scrambled control shRNA, the injections of recombinant AAV8-shRNA-TFEB reduced the expression of TFEB, as well as that of its target genes. Western blotting revealed that the knockdown of TFEB downregulated the expression of LC3B-II and upregulated the protein expression of p62/SQSTM1 (Figure 2C). Likewise, a downregulated TFEB level did not affect the protein level of Atg7. Taken together, these results confirm that metformin induces autophagy through the activation of TFEB.
In this study, Ubigene provided the recombinant AAV8 shRNA TFEB and the control AAV to study whether TFEB was involved in metformin induced autophagy. The AAV particles provided by Ubigene are ultra-purified, and the virus titer is 10^12~10^13 GC/ml, so it is highly adaptable to in vivo experiments. Click here to learn about our AAV production > >
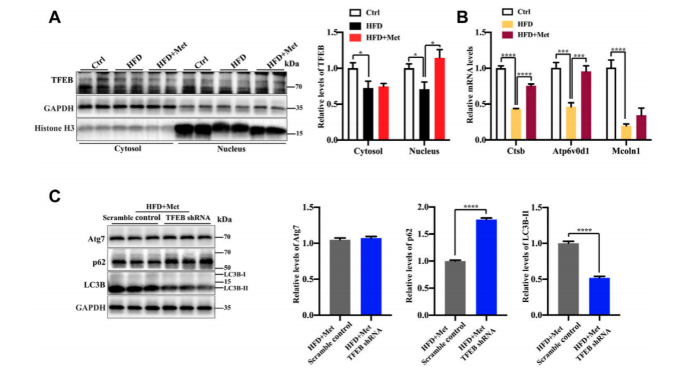
Figure 2
Metformin improves excessive lipid accumulation and IR in HFD-fed mice via TFEB
To test whether the activation of autophagy mediated by TFEB was involved in the alleviation of an excessive lipid accumulation and IR in HFD-fed mice, they measured the hepatic steatosis level using H&E and Oil red O staining after the knockdown of TFEB(Fig.3). The result showed that the protective effects of metformin on excessive lipid accumulation and IR are critically dependent on the TFEB in HFD-fed induced NAFLD mice.
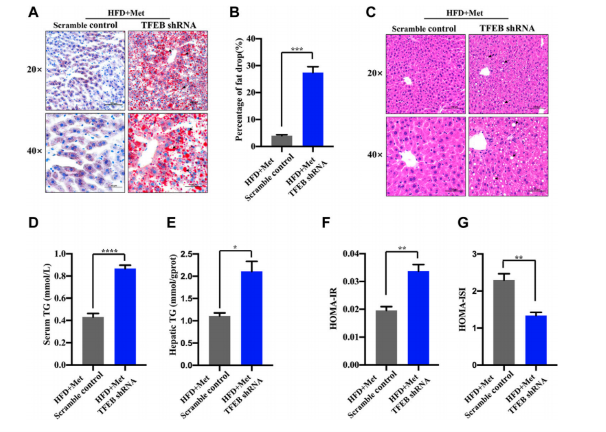
Figure 3
Overall, this study reveals a novel molecular mechanism for improving the NAFLD by using metformin, and highlight the potential beneficial effects of TFEB for the treatment of NAFLD.
Ubigene is an enterprise focusing on cell engineering. We provide high-quality gene-edited cell lines, stable cell lines, virus packaging and other related services around the world. Recentlu, Ubigene launched a KO cell bank with nearly 2000 KO cell lines. Welcome to contact our expert team to discuss your potential projects!
More article you may interested:
How U-87 MG facilitates brain tumor research?
Study the Nobel Prize with Nobel Prize: CRISPR mediated humanized MC38 cell line