Scientists have developed a continuous knockout cell line technology based on the CRISPR-Cas9 system, including single and multiple (up to three) target gene insertions and knockouts for the purpose of bacterial modification. The CRISPR/Cas system is a kind of prokaryotic immunity. The resistance of the system to foreign genetic elements (such as those found in plasmids and phages) can provide some form of acquired immunity. RNA with spacer sequence can help Cas protein recognize and cleave foreign DNA. Other Cas proteins guided by RNA can cleave foreign RNA. CRISPR is found in approximately 40% of bacterial genome sequences and 90% of archaeal sequences.
Multi-gene editing in the E. coli genome through the CRISPR-Cas9 system
The construction of industrially useful microorganisms requires effective genome-scale editing tools. The researchers describe a targeted, continuous multi-gene editing strategy that uses the Streptococcus pyogenes type II CRISPR-Cas9 system to be applied to the E. coli genome to achieve a variety of precise genome modifications, including gene deletion and insertion, with efficiency The maximum is 100%, and it can perform multi-gene editing on three targets at the same time. The system also proved that it successfully achieved targeted chromosome deletion in another Enterobacteriaceae-Tatumella citrea, with an efficiency of up to 100%.
Multi-stage knockout of Escherichia coli using CRISPR/Cas9
With the recent use of CRISPR/Cas9 technology as a standard tool for genome editing, laboratories around the world have experienced one of the largest molecular biology advances since PCR. The main advantage of this method is its simplicity and versatility for any category. Of particular interest is the widely studied Gram-negative bacterium Escherichia coli, because it is considered a major force for research and industrial applications. The researchers proposed a simple, reliable and effective scheme using the CRISPR/Cas9 system combined with the lambda Red machine for gene knockout in E. coli. In our procedure, it is crucial to use double-stranded donor DNA and a solidification strategy to remove the RNA-encoding guide plasmid, which allows the initiation of new mutations in only two working days. Our protocol allows multiple stepwise knockout strains with high mutagenesis efficiency to be suitable for high-throughput methods.
Multi-functional genome-wide CRISPR system for high throughput genotype–phenotype mapping
Genome-scale engineering is an indispensable tool to understand genome functions due to our limited knowledge of cellular networks. Unfortunately, most existing methods for genome-wide genotype–phenotype mapping are limited to a single mode of genomic alteration, i.e. overexpression, repression, or deletion. Researcher report a multi-functional genome-wide CRISPR (MAGIC) system to precisely control the expression level of defined genes to desired levels throughout the whole genome. By combining the tri-functional CRISPR system and array-synthesized oligo pools, MAGIC is used to create, to the best of our knowledge, one of the most comprehensive and diversified genomic libraries in yeast ever reported. The power of MAGIC is demonstrated by the identification of previously uncharacterized genetic determinants of complex phenotypes, particularly those having synergistic interactions when perturbed to different expression levels. MAGIC represents a powerful synthetic biology tool to investigate fundamental biological questions as well as engineer complex phenotypes for biotechnological applications.
Ubigene developed CRISPR-B™ which optimizes the microbial gene-editing vectors and process. The efficiency and accuracy are much higher than traditional methods. CRISPR-B™ can be used in gene editing of bacteria and fungi
Reference
Endo M, Mikami M, Toki S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015;56(1):41-47. doi:10.1093/pcp/pcu154
König, Enrico & Zerbini, Francesca & Zanella, Ilaria & Fraccascia, Davide & Grandi, Guido. (2018). Multiple Stepwise Gene Knockout Using CRISPR/Cas9 in Escherichia coli. BIO-PROTOCOL. 7. 10.21769/BioProtoc.2688.
Jiazhang Lian, Carl Schultz, Mingfeng Cao, Mohammad HamediRad,Huimin Zhao.Multi-functional genome-wide CRISPR system for high throughput genotype–phenotype mapping.Nature Communications .2019.
The efficiency of gene knock-out and cleavage can not only give people the ability to generate protein radical profiles and establish regulatory records, but also has many advantages, making it a particularly attractive recombinant protein expression system. First, it is carboxylated on glutamic acid and sulfated on tyrosine. Second, the operation is simple, and the recombinant protein can be quickly produced through transient gene expression. Third, it can be used for stable recombinant protein production. Some researchers used gene cell knockout and cutting efficiency systems to generate gene-edited cell lines, targeted sequencing of GLUL genomic loci, produced stable EPO cell lines, and discovered the mechanism of stable expression of recombinant erythropoietin in humans .
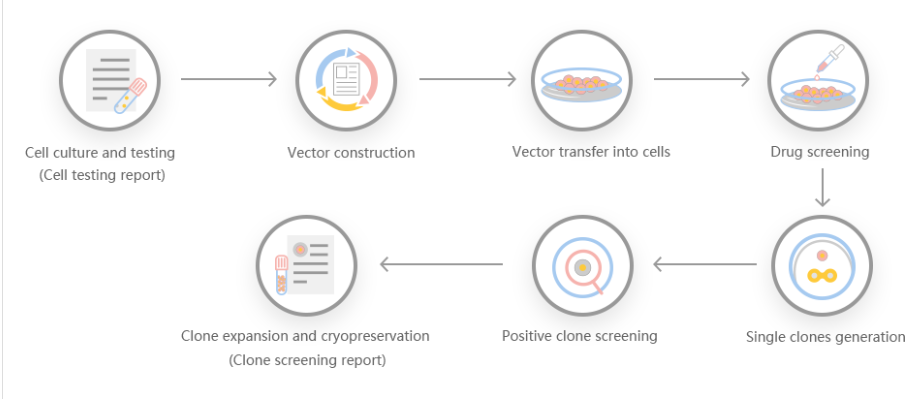
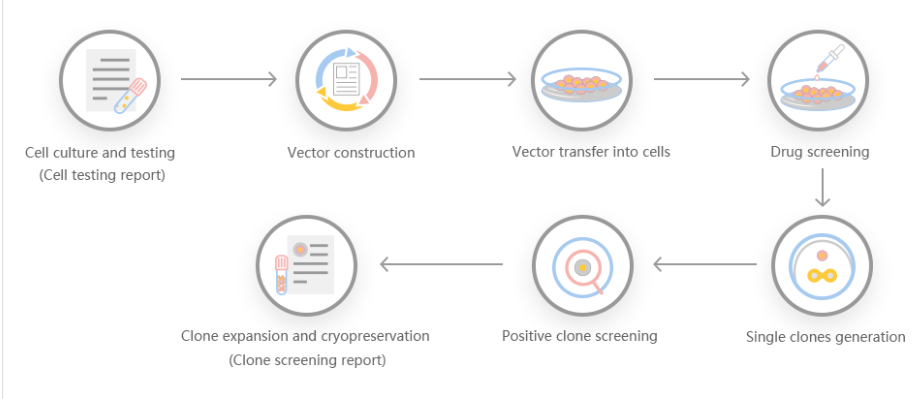
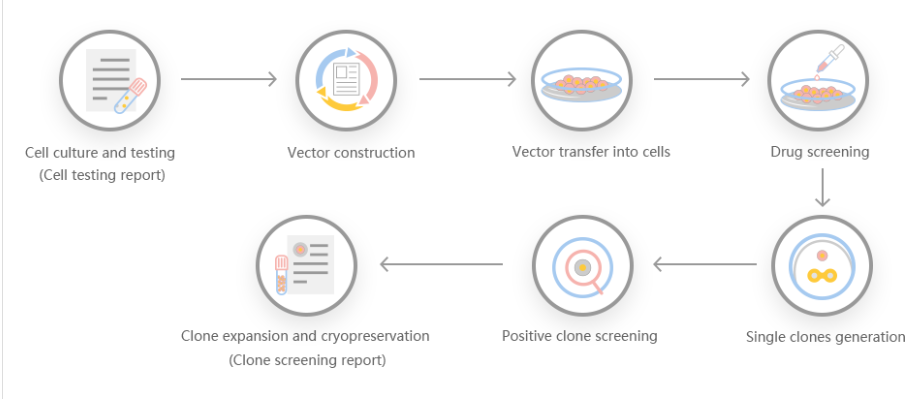
According to customer needs, Yuanjing Biotechnology designs a stable gene transfer knockout program based on the target gene.
Scheme 1: Small-segment gene knockout program, gRNA is set in the introns at both ends of exon 2, and the number of bases encoded by the knockout exon is not 3 times, and the knockout can cause frameshift.
Scheme 2: Frameshift gene knockout scheme, gRNA is set on the exon, the number of missing bases is not 3 times, and frameshift mutation can occur after knockout.
Scheme 3: Large-segment gene knockout scheme, knock out the coding sequence of the entire gene to achieve the effect of large-segment knockout.
2. Provides different virus packaging services, including lentiviruses, adenoviruses and adeno-associated viruses.3. Provides high-quality services for gene knockout, point mutation and knockin cell lines.
2. Provides culture strategies and related products for different cell types.3. Provides cell biology-related services such as cell isolation, extraction and validation.