Wednesday, July 28, 2021
Gene Point Mutation/Knock-in Cell Lineas low as $8480 | Ubigene
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Thursday, July 22, 2021
Ubigene Lentivirus Packaging/Stable Cell Line service as low as $559, 500 USD off on every 5K spent
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Monday, July 19, 2021
Red Cotton™ Gene Knockout Project | Ubigene
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Friday, July 16, 2021
Well-known SW480 cell line is not “simple” | Ubigene

Background
SW480 cell line was isolated from a 50-year-old male patient with in-situ rectal adenocarcinoma. SW480 cell line was passaged to the 91st generation when A·Leibovitz submitted it to ATCC. So far, SW480 cell line is a relatively complete cell characterization strain in many colon cancer cell lines, and its biological and genetic characteristics have been reported in many literatures. SW480 cell line belongs to adherent culture and presents epithelioid (Fig. 1), it has the following features:
(1)Carcinoembryonic antigen is positive
(2)Keratin immunoperoxidase staining is positive
(3)G of 273 codon of p53 gene → Arg is caused by A mutation →His replacement, C of 309 codon → T mutation leads to Pro → Ser replacement;
(4)The expression of oncogenes ( c-Myc, Kras, Hras, Nras, Myb, Sis, and Fos) were positive. The 12 codon missense mutation of Kras oncogene can be used as a positive control for PCR detection;
(5)No expression of cytolytic enzyme.
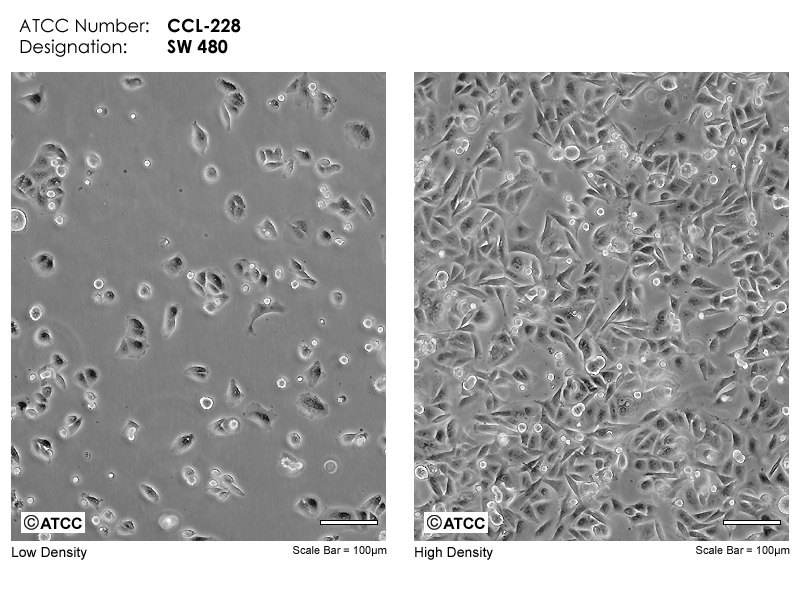
Figure 1
Reference: https://www.atcc.org/products/ccl-228#detailed-product-information
Applications and Prospect
Colorectal cancer (CRC) is one of the most common digestive system malignant tumors. The incidence rate of colorectal cancer places third in malignant tumors, and it has the second-highest mortality rate. According to GLOBOCAN data, there are 1.85 million new cases of colorectal cancer in 2018, and about 880,000 people died, which brought great threat to human health. Both Kras mutation and P53 mutation carried by SW480 cell line are pathogenic mutations with high frequency. Therefore, SW480 cell line can be used as a typical colon cancer cell model for studying the carcinogenic molecular mechanism of Kras and P53 mutations and establish drug screening platform. In addition, it can be used as a positive control for PCR identification of these mutations.
Detailed Applications
It is used to establish 3D culture model to study the mechanism of tumorigenesis
miRNAs are widely involved in tumorigenesis and biological regulation of tumor stem cell lines, and they are potential therapeutic targets. Masazumi Sakaguchi et al. used SW480 cell line in 3D culture to simulate the growth of colon cancer cells, and used this model to test the effect of miR-137 on tumor growth. This study found that miR-137 regulates the growth of tumor cells by targeting DCLK1, which reveals that miR-137 may become a drug target.(Fig.2)
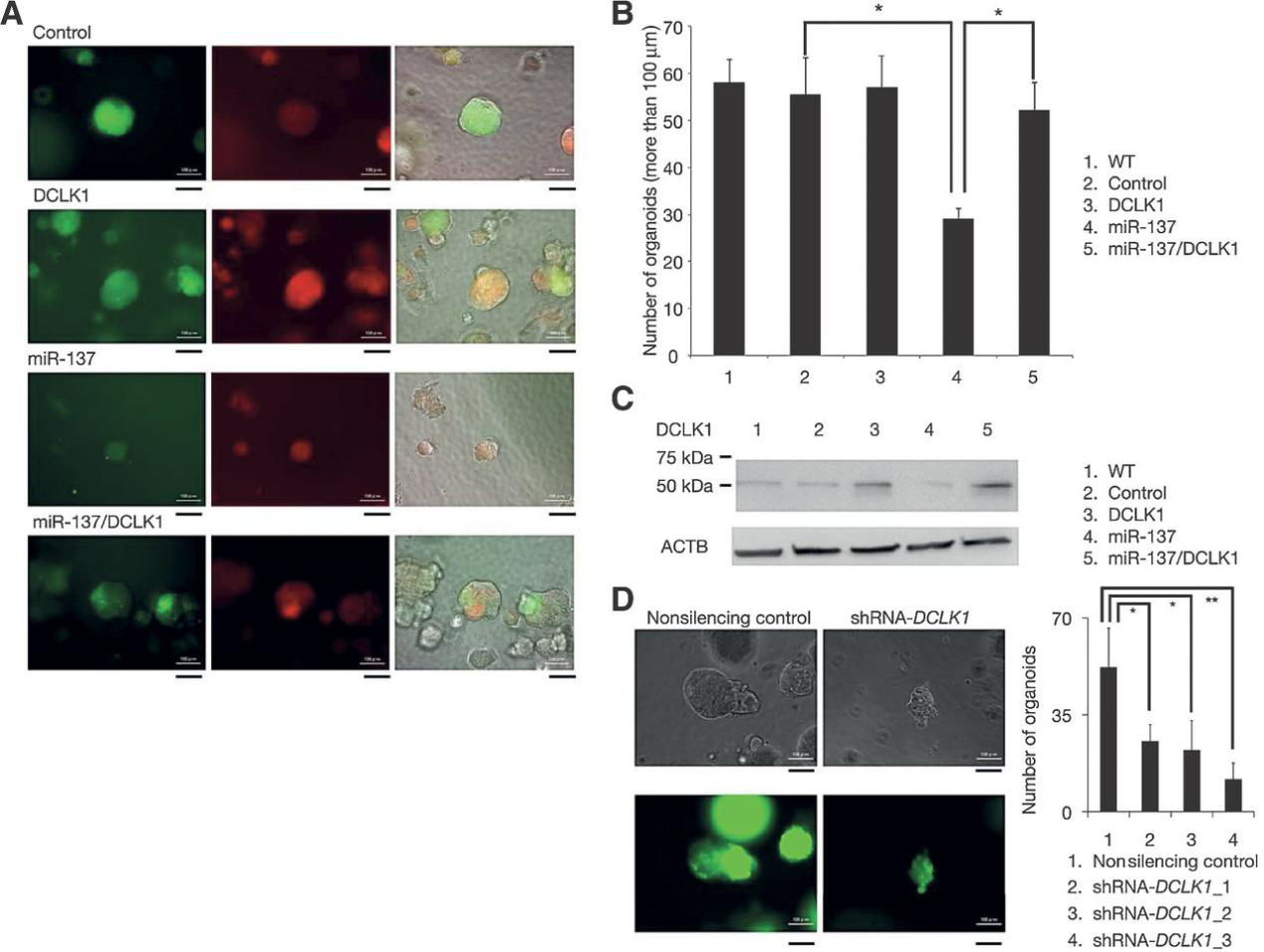
Figure 2
Biomarkers for potential tumor diagnosis:
Omentin-1 is a protein associated with colorectal cancer. Epidemiological and clinical studies have shown that the level of omentin-1 in blood is significantly increased in patients with CRC. However, the reason for the up regulation of Omentin-1 and whether it is expressed in colorectal cancer cells have not been determined. Yaqin Zhang et al. used SW480 cell line and HCT116 cell line as colorectal cancer models to explore this problem at the transcription, translation and secretion levels. The results showed that Omnitin-1 was up-regulated in tumor cells and secreted to extracellular (Fig. 3).
Figure 3
Study the effect of Kras mutation on Correlation and potential strategy of drug resistance in tumor
In colorectal cancer, the activation of RAS-RAF and PIK3CA-AKT pathway is related to the drug resistance of EGFR mAbs. In order to study the molecular mechanism of drug resistance, Stefania Napolitano et al. used several colorectal cancer cell lines with Kras mutation, such as SW480O, LOVO, HCT116, to explore the antitumor effect of related drugs. The results showed that cetuximab combined with regorafenib could overcome the resistance of anti EGFR antibody (Fig. 4).
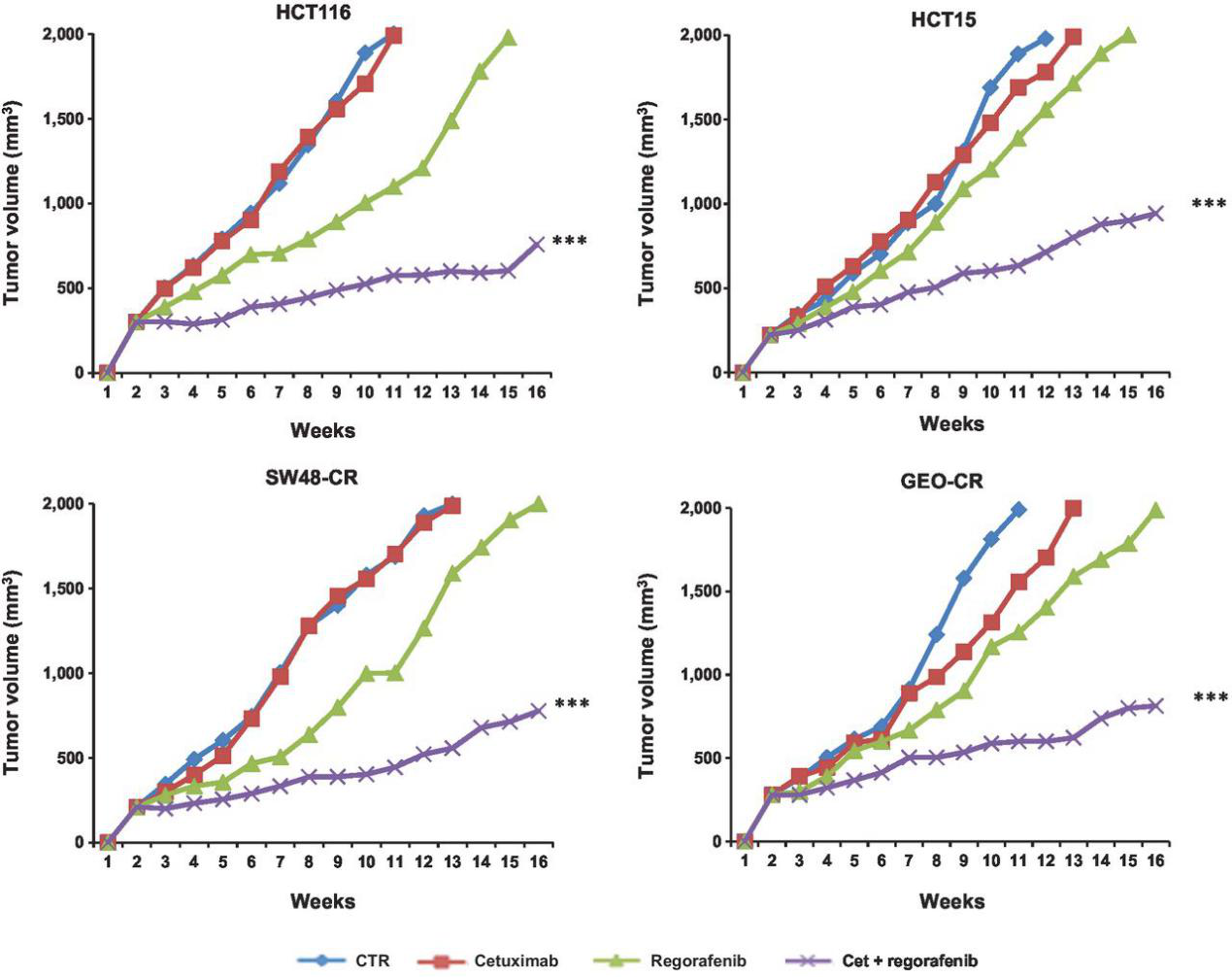
Figure 4
Case study of CRISPR/Cas9 mediated SW480 modification
CRISPR/Cas9 has been widely used in disease modeling, drug target intervention and treatment development. A number of studies have used CRISPR/Cas9 to modify the gene in SW480 cell line and other colorectal cancer cell lines to explore the biological functions and potential intervention targets of Ras, P53 and other signaling pathways in the development of colorectal cancer.
Ubigene has successfully generated KO SW480 cell line for many customers, and guarantees that 100% refund if there is protein residue. Now Ubigene has time-limited discount of gene KO cell line service, as low as 3780 USD. There is only a week left. Click here for more information >
To study the role of m6A modification of RNA in colorectal cancer, METTL3 knockout SW480 cell line is generated.
Jihao Xu et al. generated METTL3 knockout SW480 cell line with CRISPR/Cas9 technology, and found that METTL3 knockout resulted in the slowing down of SOCS2 mRNA degradation rate and up-regulation of its expression, this results to a significant inhibition of cell proliferation.(Fig.5) The study well exemplified the m6A modification of RNA as an important participant in the tumorigenic process of colorectal cancer.
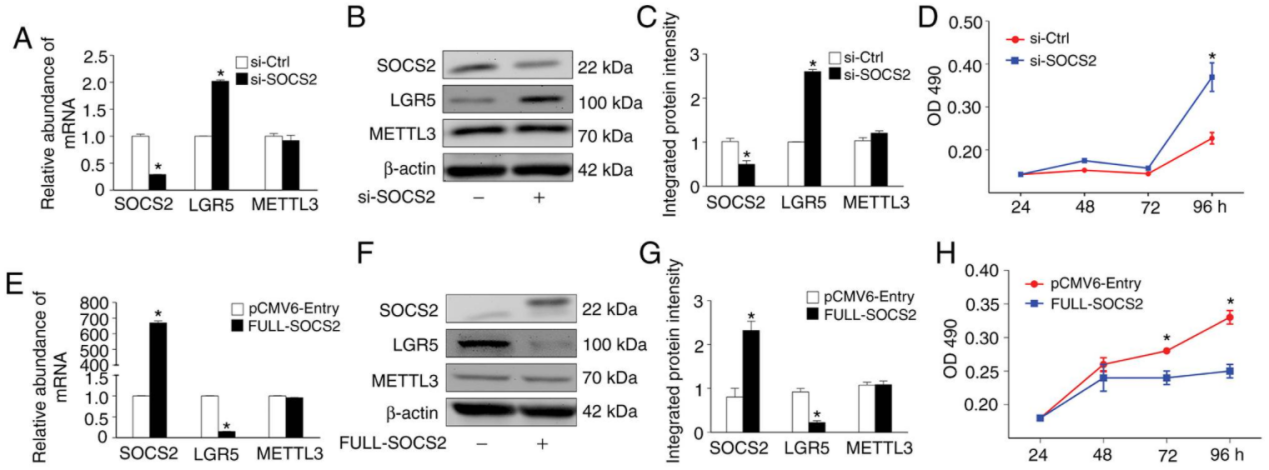
Figure 5
CRISPR/Cas9 technology introduces SNP rs6854845G>T mutation into SW480 , to explore the effect of the mutation on chromosome and gene expression:
Single nucleotide polymorphisms (SNPs) are closely associated with diseases, including cancer. Zhu Cong et al. used CRISPR/Cas9 technology to introduce SNP rs6854845 G>T mutation in primary colon epithelial cell lines, SW480 cell line and HCT116 cell line, and detected the effect of this mutation on chromosome high-level structure and gene expression. The results showed that this mutation only affected the advanced chromatin structure and gene expression of normal colonic epithelial cell lines, which was tolerated by SW480 and HCT116 colonic cancer cell lines. It is suggested that carcinogenesis could escape the regulation of chromatin conformation and gene expression by normal polymorphism.(Fig.6)
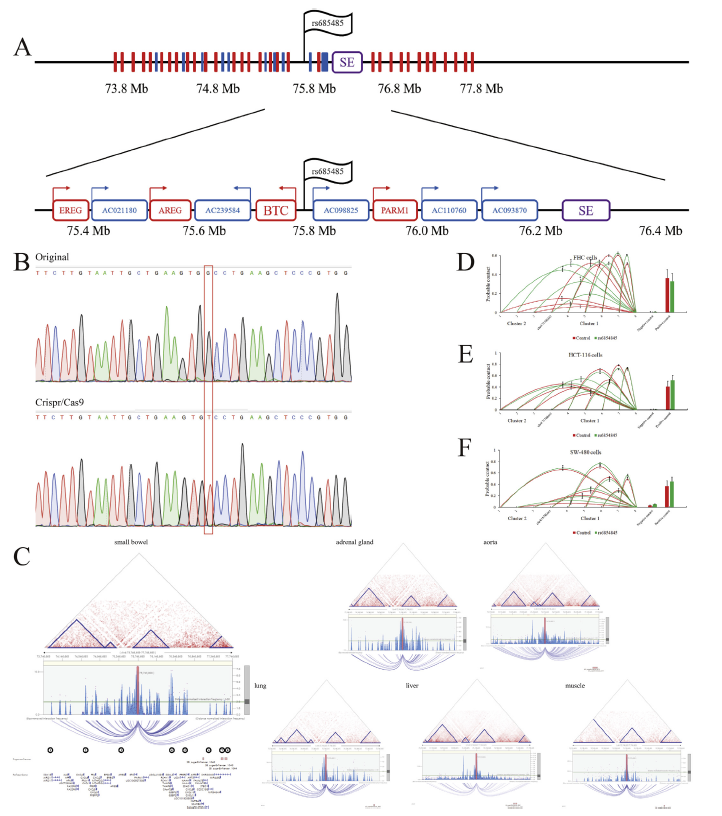
Figure 6
<
CRISPR/Cas9 technology has played a key role in the researches of cancer treatment. As a new gene-editing tool, it does have unique advantages. Therefore, Ubigene independently developed CRISPR-U™ Technology, which is 10-20 times more efficient than traditional gene editing.
So far, Ubigene has used CRISPR-U ™ technology to successfully modified genes from more than 100 cell lines, including the SW480 cell line. And Ubigene provides gene-editing services worldwide. Now Ubigene has time-limited discount of all gene-editing services: KO cell line service, as low as 3780 USD、KI/Point mutation cell line service, as low as 8480 USD、 gRNA plasmids, as low as 80 USD
Last week to join us, enjoy this great deal >;
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Wednesday, July 7, 2021
Gene editing in BEAS-2B cell line facilitates respiratory researches | Ubigene Biosciences
1.Background
Human bronchial epithelium cell line (BEAS-2B) was derived from normal bronchial epithelium obtained from the autopsy of non-cancerous individuals. This cell line comes from the ATCC CRL-9609, it is also known as bronchial epithelial cell line. The BEAS-2B cell line was initially infected by an adenovirus 12-SV40 hybrid virus, and the immortalized cell line of BEAS-2B established by continuous cell passage. BEAS-2B cell retains the ability to differentiate from serum response, this ability can be used to screen chemical or biological agents that induce or affect differentiation and carcinogenesis. Therefore, BEAS-2B cells are the ideal cell model for the study of drug metabolic activation and respiratory tumors and molecular mechanisms.
2.Applications of BEAS-2B cell line
2.1 The BEAS-2B cells shared a similar potential with hMSCs on both osteogenic and adipogenic differentiation
After culturing for 21 days under each differentiation induction condition, Oil Red O was used to stain lipid vacuoles in the differentiated adipocytes and Alizarin Red S was used to stain calcium deposits in the differentiated osteocytes. The result shows that both BEAS-2B and hMSC1 cells showed an almost equivalent positive staining for either osteogenesis or adipogenesis. These results indicate that, similar to hMSC1, BEAS-2B exhibited a strong capability of differentiating into osteocytes and adipocytes after each differentiation induction .
2.2 The BEAS-2B can be used to screen chemical and biological agents.
BEAS-2B has been extensively used to study cellular and molecular mechanisms involved in lung carcinogenesis, including the role of epithelial-mesenchymal transition (EMT) in lung carcinogenesis, as well as pneumococcal infections. In addition, the BEAS-2B cell line has been utilized as an in vitro cell model for assaying or screening various chemicals and biological agents with potential pulmonary toxicity or lung carcinogenicity.
3.CRISPR-U ™ mediated Gene editing in BEAS-2B cell line
Genome editing and the creation of cell models can advance researches in the area of functional genomics, signaling pathways, metabolism, cell death, drug discovery, drug response, and cancer research, etc. BEAS-2B is the ideal cell model for the study of drug metabolic activation and respiratory tumors and molecular mechanisms. So far, CRISPR/Cas9 technology has been used by many researchers to study cancer characteristics, disclosure of drug resistance mechanism, cancer treatment, cell death research, functional genomics, signaling pathway, drug discovery, drug response, and cell therapy, etc. CRISPR-U™ (based on CRISPR/Cas9 technology), developed by Ubigene, is more efficient than general CRISPR/Cas9 in double-strand breaking, and CRISPR-U™ can greatly improve the efficiency of homologous recombination, easily achieve gene editing in vitro and in vivo. It has great advantages for gene editing of BEAS-2B cells.
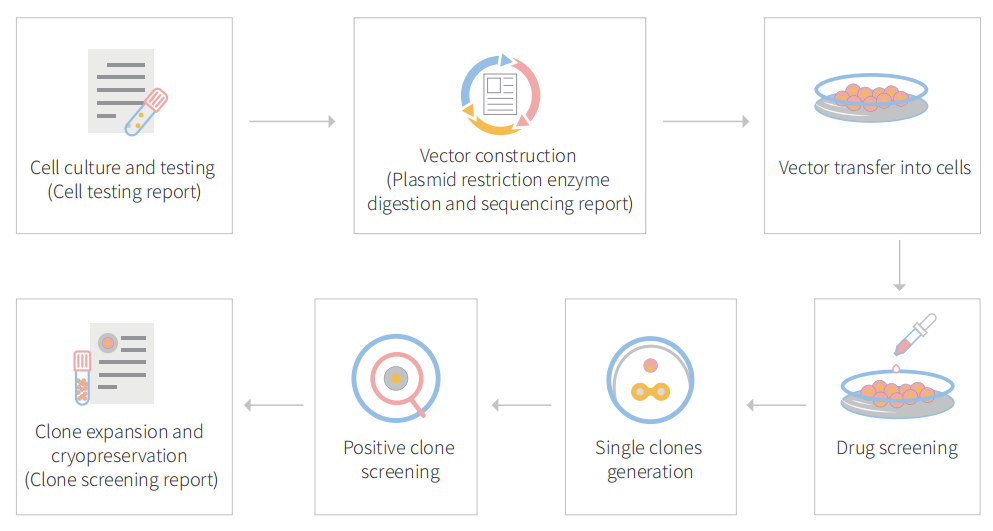
Figure 1 Gene editing in BEAS-2B cells with CRISPR-U™
3.1 Gene knockout in BEAS-2B cell line with CRISPR-U™
gRNA and Cas9 would be transferred into cells by virus transduction, liposome transfection, or nucleofection. After drug screening, single clones would be isolated. Positive clones would be validated by sequencing. Specific case is as shown below:
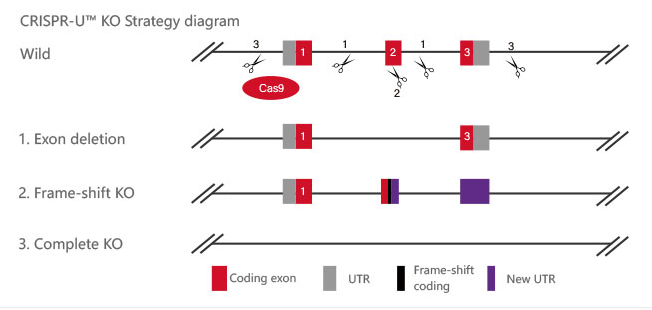
Figure 2 Knockout Strategies of BEAS-2B cell line
P53 gene is an important factor in the malignant transformation of BEAS-2B cells by radon. The P53 gene was knocked out in human normal lung epithelial cells (BEAS-2B) and radon was treated for gene knockout BEAS-2B cells and normal BEAS-2B cells, respectively. The researchers found earlier malignant transformation of BEAS-2B cells from radon after knocking the P53 gene. The BEAS-2B cell (P53 KO-Rn) proliferate faster (BEAS-2B <0.05), earlier cloning (P <0.05), more invasive (P <0.05), and earlier changes in mRNA expression levels of epithelial-interstitial transformation (EMT) -related genes (P <0.05). The results show that radon irradiation leads to malignant transformation of BEAS-2B cells and that P53 plays an important role in radon gas-induced lung cancer.
Reference :http://www.paper.edu.cn/releasepaper/content/201910-45
3.2 Gene KI/point mutation BEAS-2B cell line
BEAS-2B cells would be co-transfected with gRNA, Cas9 and donor vector by electroporation, and then screened. After drug screening, single clones would be generated. Positive clones would be validated by sequencing. Specific cases are as shown below:
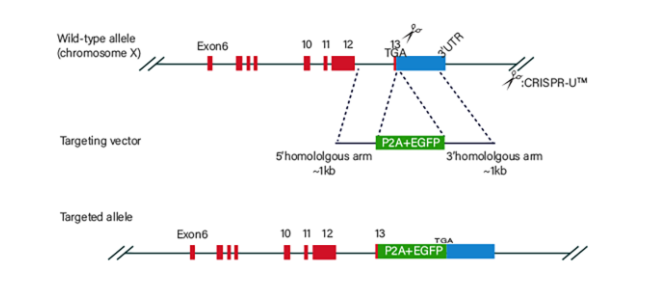
Figure 3 Knock-in Strategies of BEAS-2B cell line
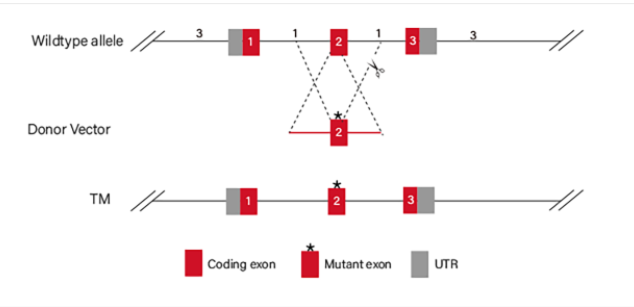
Figure 4 Point mutation Strategies of BEAS-2B cell line
(1)Case of point mutation cell line-1
To study the specific influence of NRASQ61K mutant on angiogenesis, NRASQ61K was knocked into normal (non-neoplastic) human bronchial epithelial cells (BEAS-2B). Compared to wild type (WT) cells, NRASQ61K expressing cells growing on CAM were characterized by a disorganized vasculature with several hemorrhagic areas. To confirm the proangiogenic effects of NRASQ61K in vivo, a Matrigel plug assay was performed in athymic nude mice. As shown in Figure 5B, NRASQ61K -expressing cells demonstrated a marked increase in angiogenesis in the plugs, as indicated by a bright red color. However, angiogenesis was negligible and hemoglobin levels were lower in the WT cells. As shown in Figure 5C, cells spontaneously form capillary-like structures on Matrigel, which is a critical step in tumor angiogenesis. Therefore, NRASQ61K expression has the effect on tubulogenesis.
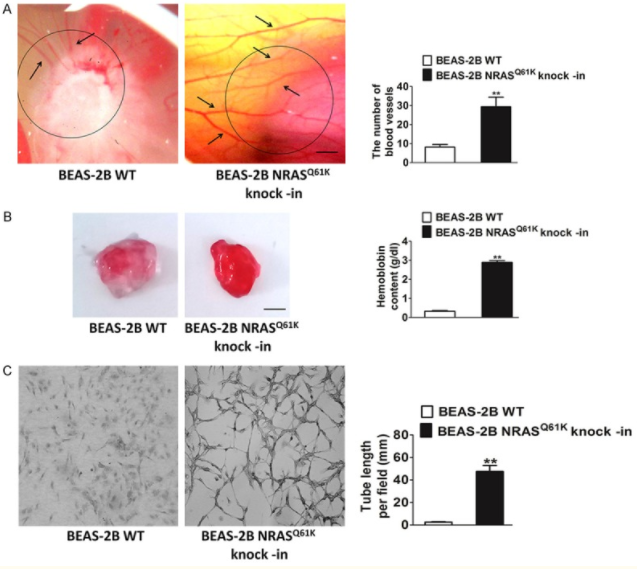
Figure 5 Knock-in of NRASQ61K into BEAS-2B
(2)Case of point mutation cell line-2
Arsenite Exposure Led to Activation of NFAT but Not AP-1 and NFκB in Beas-2B Cells, the COX-2 gene promoter region contains NFκB, AP-1, and NFAT binding sites, which can be recognized by these transcription factors and in turn lead to COX-2 transcription. To determine whether NFAT regulated COX-2 expression through direct binding to the COX-2 promoter region, point mutation of the two NFAT binding sites in the promoter region of COX-2-Luc reporter was carried out by CRISPR/Cas9. The results demonstrated that this mutation resulted in impairment of the COX-2 transcription induced by arsenite exposure. And the researchers found that it may be noted that NFAT activation by arsenite in the time course studies reached a peak earlier, as compared with that of COX-2 induction. Because the exposure of arsenite only causes NFAT activation in BEAS-2B cells, not AP-1 and NFκB activation. Thus, the activation of NFAT by arsenite is the reason for COX-2 induction in BEAS-2B cells.
In addition, to confirm the important role of NFAT3 in arsenite-induced COX-2 expression in BEAS-2B cells, the researchers built and used the siNFAT3. They used the cells for 10 μm NFAT selective inhibitors were pretreated and then exposed to 20μm arsenite. The results demonstrated that stable transfection of siNFAT3 in Beas-2B cells resulted in a dramatic reduction of NFAT3 protein expression in Beas-2B cells (Fig. 6b). Specific knockdown of NFAT3 expression by siNFAT3 blocked arsenite-induced COX-2 transcription and protein expression (Fig. 6, e and h).
These results distinctly demonstrate that NFAT3 is a major mediator for COX-2 induction by arsenite in Beas-2B cells.
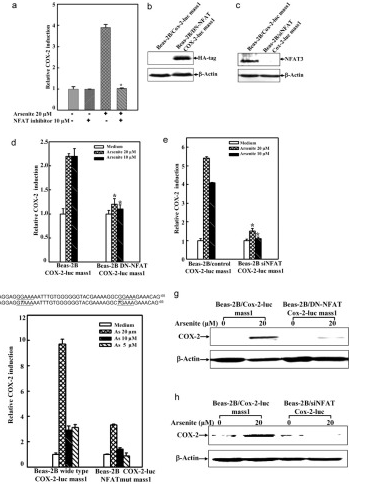
Figure 6 Point mutation cases of BEAS-2B cell line
Ubigene’s CRISPR-U ™ technology and more than 5000 successful gene-editing cases make our services more trustworthy! Now we have time-limited discount of all gene-editing services: KO cell line service, as low as 3780 USD、 KI/Point mutation cell line service, as low as 8480 USD、 gRNA plasmids, as low as 80 USD
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Sunday, July 4, 2021
What assays you need after stable cell line generation? Ubigene

As long as we construct gene editing cell lines (KO, KI, point mutation and stable expression), cell phenotype needs to be identified. However, there are so many ways to test cell phenotypes, which one is the best one to fullfill our research goal?
Most of the cell line related studies will perform cell assays of proliferation, cytotoxicity, cell cycle, cell migration, invasion and apoptosis. We may know the answer by understanding the principle and application of these assays!
Cell proliferation is the basis of growth, development, reproduction and heredity of cells. By comparing the growth curves of different groups, such as wild-type vs mutant, control vs overexpression, control vs knockdown and so on, we can identify whether there are significant differences in the proliferation status of the constructed cell lines, and then we can choose to detect the cell cycle or apoptosis. Figure 1 shows the effect of p62 overexpression on U87 and U251 proliferation.
There are many methods to detect cell proliferation, including CCK-8 detection and Trypan Blue Cell Counting. Cell counting is a necessary skill for researchers. CCK-8 detection method is based on the principle that succinate dehydrogenase in mitochondria of normal cells can reduce tetrazolium salts to purple crystalline substances. When these purple substances are deposited around the cells, cell proliferation status can be detected under 450nm OD (optical density) by microplate reader. Other methods, such as BrdU staining and Ki-67 detection, can reflect the spatial distribution of proliferating cells. However, these methods can only capture the proportion of proliferating cells at a single time point, so they are generally used to test cell proliferation in tissue sections.

Figure 1
Reference : Deng, Danni et al. “p62 acts as an oncogene and is targeted by miR-124-3p in glioma.” Cancer cell
international vol. 19 280. 6 Nov. 2019, doi:10.1186/s12935-019-1004-x
Generally, regarding the changes of cell proliferation, by detecting the cell cycle, we can find out the reason of the changes. For example, the number of cells in S, G2 or M phase decreased, but most of them were arrested in G1 phase, which shows that G1/S checkpoint may be abnormal. This is of great significance for further understanding of biological growth and control of tumor growth. Propidium iodide (PI) staining flow cytometry is a simple and direct method to detect cell cycle.
Propidium iodide (PI) can emit fluorescence when combined with dsDNA, and the fluorescence intensity is proportional to the content of double-stranded DNA. Therefore, the DNA content of cells can be determined by flow cytometry. Because the DNA content of each phase of cell cycle is different, each phase of cell cycle can be divided into G1/G0 phase, S phase and G2/M phase according to the distribution of DNA content. The percentage of each phase corresponding to the flow cytometry histogram can be calculated by software (Fig. 2), and then the cell cycle can be analyzed.
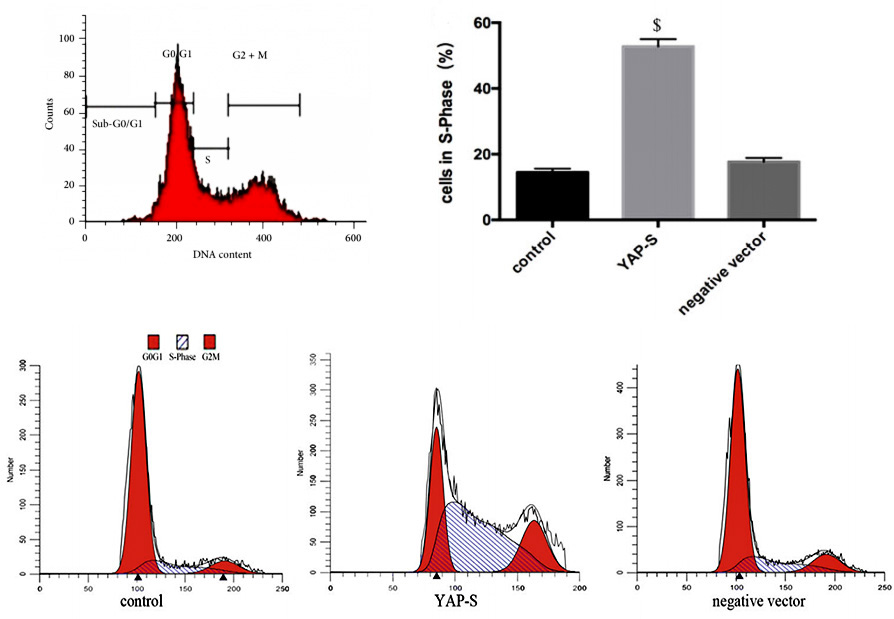
Figure 2
Reference: https://ibook.antpedia.com/z/93690.html
Xie, Kewei et al. “Yes-associated protein regulates podocyte cell cycle re-entry and dedifferentiation in adriamycin-induced
nephropathy.” Cell death & disease vol. 10,12 915. 4 Dec. 2019, doi:10.1038/s41419-019-2139-3
Apoptosis is a process strictly controlled by multiple genes, such as Bcl-2 family, Caspase family, anti-cancer gene P53, etc. The disorder of apoptosis may be directly or indirectly related to many diseases, such as tumor and autoimmune diseases. Therefore, if you are studying tumor or degenerative disease, it is necessary to detect apoptosis, which is very important for the research.
As the process of apoptosis has typical morphological and molecular changes, detection methods are diverse, such as Annexin/PI staining protocol, TUNEL detection and Caspase 3 Activity Assay. In addition, Annexin/PI staining protocol is widely used, because it can distinguish early and late apoptosis and facilitate quantification. In the early stage of apoptosis, Phosphatidyl serine (PS) will flip from the inner side of the cell membrane to the surface of the cell membrane and be exposed to the extracellular environment. Annexin-V can specifically bind with PS. FITC, a green fluorescent molecule, can be linked to Annexin-V, and then can be used to mark the apoptotic body with intimal turnover. When apoptosis occurs, the permeability of cell membrane increases, PI can enter the cell, bind to DNA and emit fluorescence. Therefore, by analyzing the signal distribution of Annexin-V-FITC and PI, we can distinguish the early apoptotic (Annexin-V-FITC positive + PI negative) and late apoptotic (Annexin-V-FITC positive + PI positive) cells (Fig. 3).
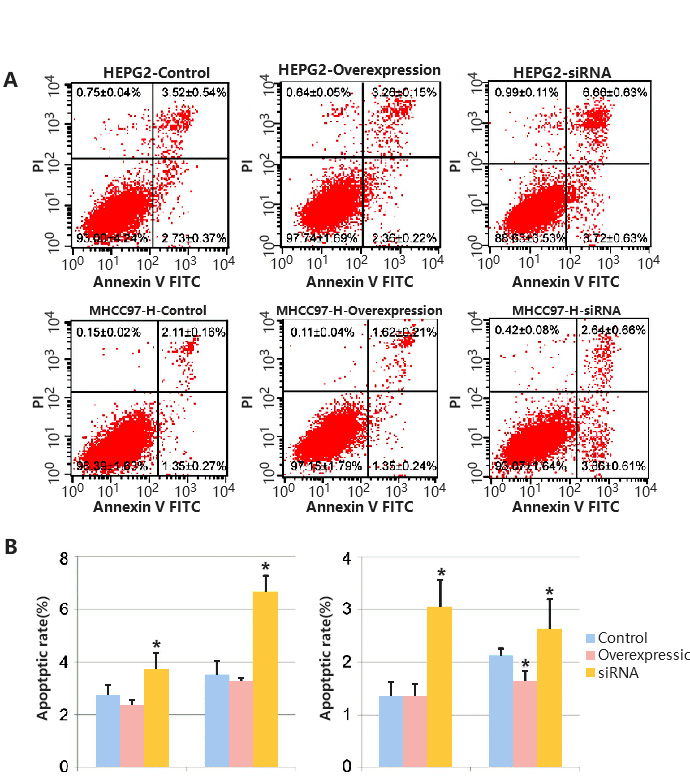
Figure 3
Reference: Chen G, Zhao X, Tan Z, et al. Investigation of the role of cullin 4A overexpression in human liver cancer.
Mol Med Rep. 2018;18(3):2531-2540. doi:10.3892/mmr.2018.9233
In the study of cell-based disease models, you may want to identify potential therapeutic drugs through high-throughput screening of in-stock drugs on the basis of overexpression, knockdown or cell line KO of potential drug targets, or it is necessary to test the cytotoxicity of specific substances, such as drug screening. Then you can use cytotoxicity tests to screen for sensitive compounds, or to screen different cell lines for sensitivity to multiple concentrations of a compound. There are many methods to achieve cytotoxicity detection, among which CCK-8 and ATP detection methods based on cell metabolic activity indicate cell viability (Fig. 4), while LDH detection method based on metabolic enzyme leakage indicates cell damage (Fig. 5). Specific methods can be selected according to the research needs.
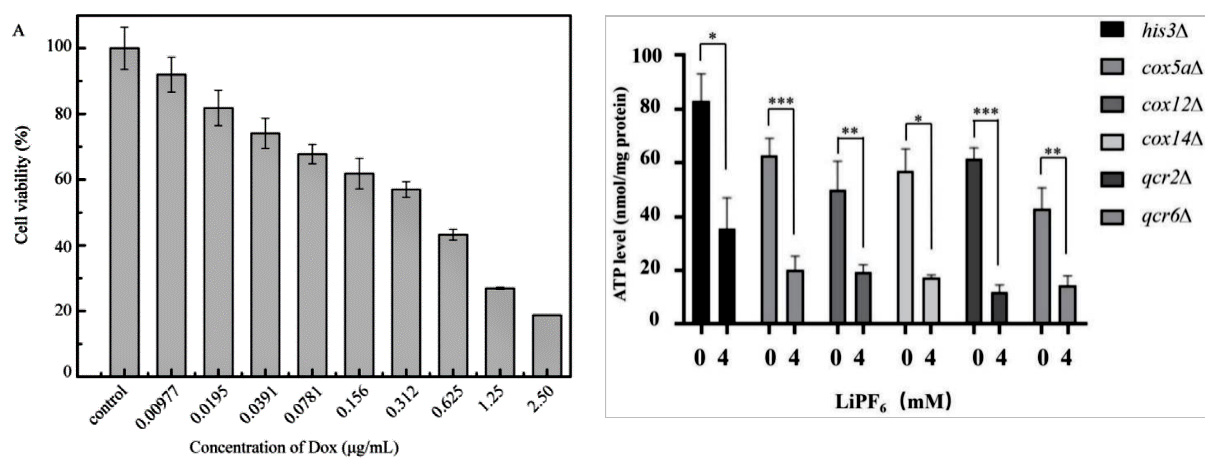
Figure 4
Reference:Cai L, Qin X, Xu Z, Song Y, Jiang H, Wu Y, Ruan H, Chen J. Comparison of Cytotoxicity Evaluation of Anticancer Drugs between Real-Time Cell Analysis and CCK-8 Method. ACS Omega. 2019 Jul 11;4(7):12036-12042. doi: 10.1021/acsomega.9b01142. PMID: 31460316; PMCID: PMC6682106. Jin X, Zhang J, An T, et al. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate. Cells. 2021;10(4):888. Published 2021 Apr 13. doi:10.3390/cells10040888
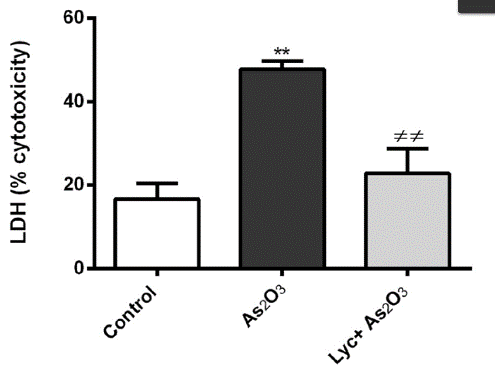
Figure 5
Reference:Oguz E, Terzioglu Bebitoglu B, Acet G, Hodzic A, Hatiboglu N, Ada S. Effect of lycopene on As2O3 induced oxidative stress in SH-SY5Y cells. Mol Biol Rep. 2021 May 4. doi: 10.1007/s11033-021-06377-y. Epub ahead of print. PMID: 33948854.
It is generally believed that inhibiting tumor cell migration and cell invasion is an effective treatment in tumor therapy. Therefore, the detection of cell migration and cell invasion ability after the intervention of tumor cells (overexpression, knockdown, knockout, point mutation, screening assay) can be used as an effective indicator to evaluate the effect of intervention.
Transwell test is commonly used to detect cell migration and invasion (Fig. 6). Transwell test is also known as "perforation experiment". The chamber is placed in a 24-well plate with complete medium. And it contains dense pores, and the cell suspension is added into the chamber. Cells can deform through the holes in the chamber and migrate to a more nutritious chamber and stick to the outer side. Finally, the cells outside the original chamber were counted by staining, then we can evaluate the cell migration and invasion ability of the cells. However, there are still differences in the experimental methods for detecting cell migration and invasion. Invasion detection needs to lay a layer of matrix gel on the polycarbonate membrane to imitate the extracellular matrix.
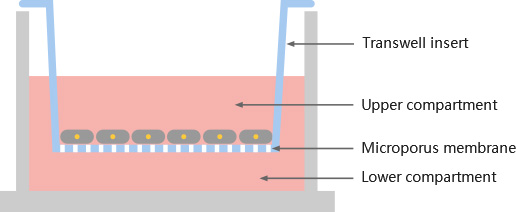
Figure 6
Reference:https://www.corning.com/worldwide/en.html
In most cases, several assays need to be performed to bring out a conclusion of how the modification affects the cell phenotype. You may have some questions: How to do three or four assays at the same time? Is it necessary to go from cell proliferation test to cell migration / invasion test? How to deal with too many experimental groups?
What can you do, of course, is to find Ubigene! We provide gene-editing cell services, as well as related plasmids and cell-related assay services. Now we are having a combo of cell assays, you can get 3 professional test and analysis reports, you will no longer be annoyed by data analysis! Click to learn more about our combo service
Ubigene has been trying to meet the needs of every customer. Ubigene has promoted cell-related assay services (cell proliferation, apoptosis, cell migration/invasion detection, cell cycle, toxicity detection, Marker protein expression detection, cytokine release/enzyme activity detection and other popular services) and awaited your choice!
 Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
Ubigene Biosciences, is a fast-growing company with an experienced team in gene-editing. Our advanced molecular and cell biology platform ensure us to provide high-quality services and products for biological research and biotechnology industries.
[Research highlight] Enhancing p53 pathway can efficiently suppress colon cancer
Colorectal cancer is the third most diagnosed cancer and leads to the second mortality among cancers worldwide. The first-line chemotherap...
-
A549 is a model lung cancer cell line developed in 1972 by scientist Giard, through the removal and culturing of cancerous lung tissue in ...
-
Background MC38 cell line was derived from the colon adenocarcinoma cell line of C57BL/6 mice, which is adherent growth and fibroblast mor...










