Background
Vero cell line, also known as green monkey kidney cell line, is a non-doubled renal cell line of African green monkey (belongs to Chloroebus). In 1962, Chiba University in Japan isolated the renal epithelial cells of normal adult African green monkeys and obtained the Vero cell line. Vero cell line is a continuous aneuploid cell line, which means that its chromosome number is abnormal. As a continuous cell line, Vero cell line can go through many division cycles without aging. Vero cell line has defective interferon secretion function. Unlike normal mammalian cell lines, they do not secrete interferon when infected by virus α/β. However, they still have interferon- α/β. Therefore, when recombinant interferon is added to its culture medium, they can still respond. So far, Vero cell line is widely used in the study of the molecular mechanism of virus infection and the production of vaccines and recombinant proteins, and are regarded as an ideal cell model for cultivating influenza vaccines and studying the molecular mechanism of virus infection [1]. WHO approved Vero cell line as a vaccine production cell and recommended it as an alternative matrix for influenza vaccine production.
Detailed applications
1.Vero cell line is the most commonly used cell line for the production of viral vectors and vaccines.
Vero cell line is considered to be non-tumorigenic within a limited generation, so it can be used as a matrix for vaccine research. In 2005, Vero cell line was approved by the World Health Organization as a cell line for the production of human vaccines. Vero cell line has been used to produce polio vaccine, rabies vaccine and ACAM200 smallpox vaccine [2].
2.Vero cell line can perform extensive cell characteristic research and the creation of large cell bank
After Vero cell line was isolated, several subcellular lines were derived, such as Vero81, Vero76 and VeroE6. Genomic analysis showed that these cell lines originated from a female African green monkey. Because Vero cell line can be subcultured indefinitely, it can be used for a wide range of cell characteristics and the creation of a large cell bank[3].
3.Vero cell line is widely used to study the molecular mechanism of virus infection
The insufficient expression of interferon in Vero cell line is considered to be one of the reasons why Vero cell line is very sensitive to many viruses, including simian vacuolar virus, measles virus, rubella virus, arthropod carrying virus and adenovirus. Later, it was found that it was also easy to be infected with bacterial toxins, including diphtheria toxin, Heat Intolerant enterotoxin and Shigella like toxin. They do not secrete signal peptide interferon after virus infection, so the antiviral defense mechanism of cells is damaged[3]. This characteristic makes Vero cell line widely used in virology, bacteriology, parasitology and toxicology [3].
Application of CRISPR/Cas9 technology in Vero cell line
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is the pathogen of COVID-19 (COVID-19) in 2019. It has triggered the most influential public health crisis in the 21st century. At this time, more than 7.5 million people around the world had been infected and more than 420,000 people died from infection. And the data is still growing. An effective vaccine is a powerful “weapon” for human to win this “fierce battle”, and determining the host factors necessary for infection is very important for understanding the pathogenesis of COVID-19, revealing the changes of host susceptibility and determining new host oriented therapy, which may be effective for current and future pandemic Coronavirus. CRISPR/Cas9 technology has become one of the main research methods for the precise editing of genome.
Ubigene also has successfully used CRISPR-UTM technology to modify the gene in Vero cell line. Because the gene knockout efficiency of CRISPR-UTM technology is leap to 80%, gene knockout in Vero cell line using CRISPR-UTM technology has a higher success rate than using traditional CRISPR/Cas9 technology. Vero-E6 cell line is highly sensitive to SARS-COV-2 infection and virus induced cytopathic effects. Wei, Jin et al. conducted two independent genome-wide CRISPR screening in Vero-E6 cell line and used Vero-E6 cell line of two different Cas9 nuclease constructs (Cas9-v1 and Cas9-v2). They evaluated 25 of these genes, including pro-viral and antiviral genes (Fig. 1). They found that pro-viral and antiviral genes are of great significance for understanding the pathogenesis, treatment and vaccine design of COVID-19. For example, SMARCA4 can promote the proliferation of alveolar epithelial cells and prevent pulmonary fibrosis, a common disease of SARS-CoV and SARS-CoV-2[4].
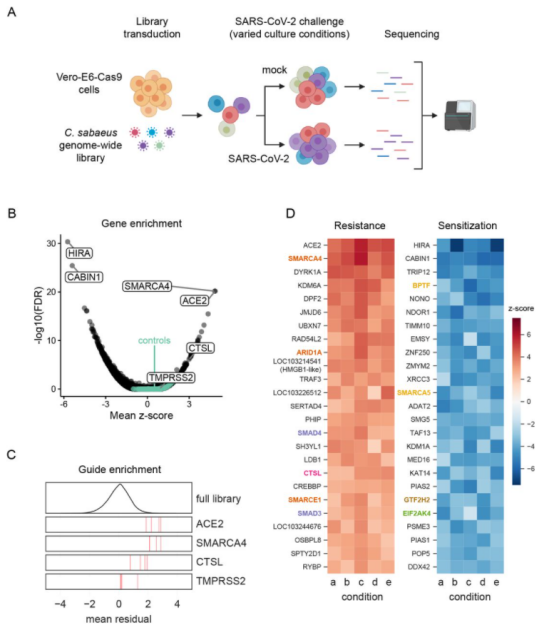
Figure 1 Genome wide CRISPR screening can identify genes crucial to SARS-CoV-2 induced cell death
Case Study
1.Study invasion and infection of Vero cell line by porcine epidemic diarrhea virus with CRISPR/Cas9 system.
Porcine Epidemic Diarrhea Virus (PEDV) is a positive single chain virus, belonging to the single chain virus genus of coronavirus subfamily. Aminopeptidase N (APN or CD13) is a type II zinc metalloproteinase, which can mediate a variety of cell physiological processes, including antigen presentation, cell differentiation, cell movement and coronavirus entry. Since porcine APN (pAPN) is the main receptor of porcine enteropathogenic α coronavirus (TGEV). Ji, Chun Miao et al. found that neither hAPN nor pAPN was the functional receptor of PEDV by knocking out the endogenous expression of hAPN or pAPN in Vero cell line. In order to completely exclude potential vAPN production in Vero cell line that may be below the detection limit, they used the CRISPR/Cas9 system to generate vAPN knockout cell line, that is, targeting vAPN-CDS1 with two gRNA/Cas9 complexes and deleting the 160 bp fragment between 290-449 nucleotides downstream of the translation initiation site (Fig. 2). These knockout cells were confirmed by IFA that Vero-vAPNKO1 and Vero-vAPNKO2 lacked the expression of vAPN (Fig. 3a). The results showed that Vero-vAPNKO1 and Vero-vAPNKO2 cell lines showed similar CPE and GFP expression to normal Vero cell line after PEDV-GFP infection, indicating that vAPN knockout had no effect on PEDV entry and infection [5].
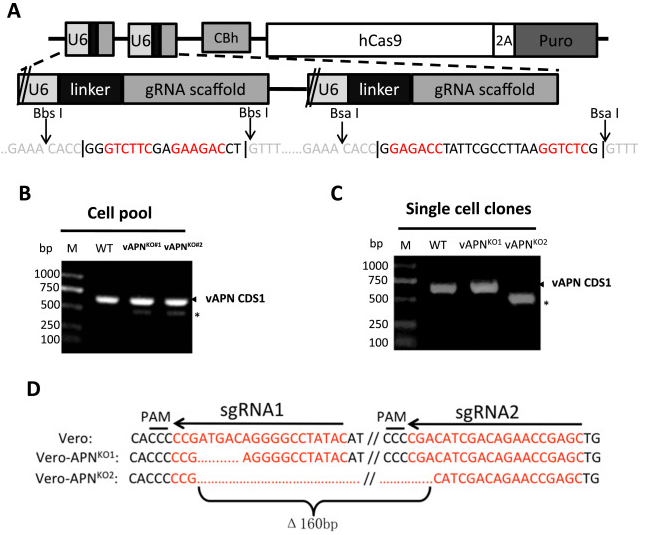
Figure 2 vAPN gene knocked out in Vero cell line via CRISPR/Cas9
Figure 3 Time resolved immunofluorescence
2.Development of enhanced RV vaccine substrates via using gene knockout Vero cell line
Rotavirus (RV) is the main cause of severe gastroenteritis worldwide. Nichole et al. used siRNAs to screen host genes that have a negative impact on RV replication, and then knocked out the selected genes through CRISPR/Cas9 gene editing. Compared with wild-type Vero cell line, fully sequenced gene knockout Vero cell line substrates can increase RV replication and RV vaccine antigen expression. The results showed that compared with other tested Vero cell line substrates, Vero cell line with EMX2 gene deletion had higher RV replication and antigen production. Gene knockout Vero cell line can promote the development of enhanced RV vaccine substrate and help to improve RV vaccine production.
Figure 4
3.CRISPR/Cas9 mediated Knock-in HSV DNA genome in Vero cell line
Bacterial Artificial Chromosomes (BACs) is the powerful tool for controlling the large genome of DNA viruses (such as herpesvirus). Because the genome of these DNA viruses (herpesviruses) is too large to be cloned into plasmids. For example, Herpes virus (HSV), a common DNA virus with a genome length of 152 kbp, may cause cheilitis, genital herpes and encephalitis. However, mutant HSV is a candidate for tumor therapy and can be used to kill tumor cells. Suenaga et al. used CRISPR/Cas9 system and Vero cell line as matrix to study mutant HSV. This method can achieve gene knockdown and gene knock-in of HSV without inserting artificial genes into the virus genome, and improve the isolation efficiency of expected mutant virus clones. In addition, CRISPR/Cas9 system can also be applied to other DNA viruses, such as Epstein Barr virus, cytomegalovirus, vaccinia virus and baculovirus. CRISPR/Cas9 system plays a very important role in the study of various types of viruses, including clinical isolated cell lines [6].
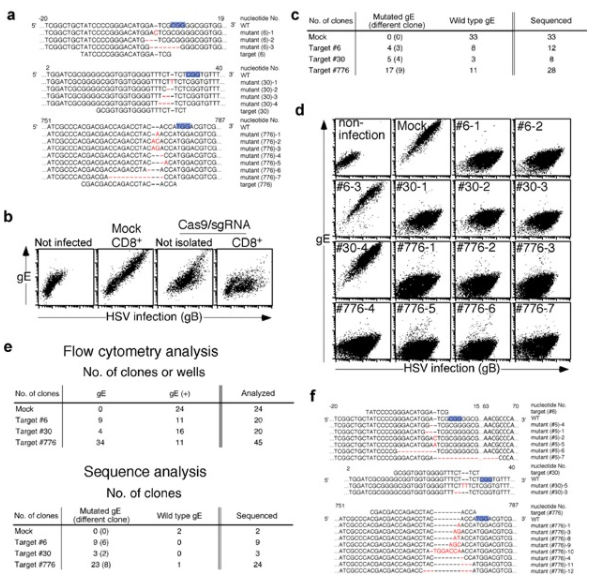
Figure 5 Gene specific mutations were introduced into HSV-1 genome by CRISPR/Cas9
So far, Ubigene has used CRISPR-UTM system to successfully modified genes from more than 100 cell lines, including the Vero cell line. And Ubigene provides gene-editing services worldwide. Now Ubigene has time-limited discount of all gene-editing services: KO cell line service, as low as 2021 USD. Ubigene provides over 600 types of in-stock KO cell lines. Over 10,000 in-stock gRNA plasmids, only 80 USD. Lentivirus Packaging, as low as 559 USD. Stable Cell Line, only 1780 USD. KI/Point mutation cell line service, as low as 8480 USD.